Bacterial Communities and Biofilms
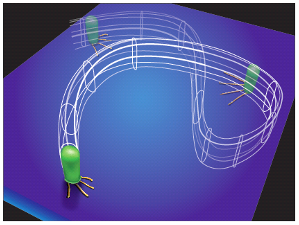
 We are reaching the limits of what can be learned using exclusively traditional
methods based on genetic manipulation and subsequent observation, since biofilm
development depends strongly on epigenetic and communal factors. We have
recently developed a general method to analyze development of bacterial biofilm
phenotypes in situ. Time-lapse microscopy movies of bacteria are translated into
searchable databases. "Search engines" are used to find in these movies all
examples of specific events in the bacterial life cycle, such as surface
attachment, cell division, or cell detachment. We can delineate the full
"geneological" and motility history for every individual cell in a movie. By
combining this high-throughput microscopy analysis with the library of mutants
available for P. aeruginosa (where 80% of the non-house keeping genes have been
mutated singly and in combination), we will be able to investigate the
connection between nutritional conditions, polysaccharide production, cdiGMP
signaling, and surface motility in P. aeruginosa biofilm formation at the
community level for the first time.
We are reaching the limits of what can be learned using exclusively traditional
methods based on genetic manipulation and subsequent observation, since biofilm
development depends strongly on epigenetic and communal factors. We have
recently developed a general method to analyze development of bacterial biofilm
phenotypes in situ. Time-lapse microscopy movies of bacteria are translated into
searchable databases. "Search engines" are used to find in these movies all
examples of specific events in the bacterial life cycle, such as surface
attachment, cell division, or cell detachment. We can delineate the full
"geneological" and motility history for every individual cell in a movie. By
combining this high-throughput microscopy analysis with the library of mutants
available for P. aeruginosa (where 80% of the non-house keeping genes have been
mutated singly and in combination), we will be able to investigate the
connection between nutritional conditions, polysaccharide production, cdiGMP
signaling, and surface motility in P. aeruginosa biofilm formation at the
community level for the first time.
We are part of the Human Frontiers Science Program Center for Early Biofilm Development.
Recent Publications
Here is some of our recent work.
How does P. aeruginosa organize into microcolonies?
The connection between M. xanthus, lubricants, and earthquakes
Why do P. aeruginosa ‘twitch’ when they crawl on a surface using their Type IV Pili?
Related Publications
More general references for our work on bacteria are listed below.